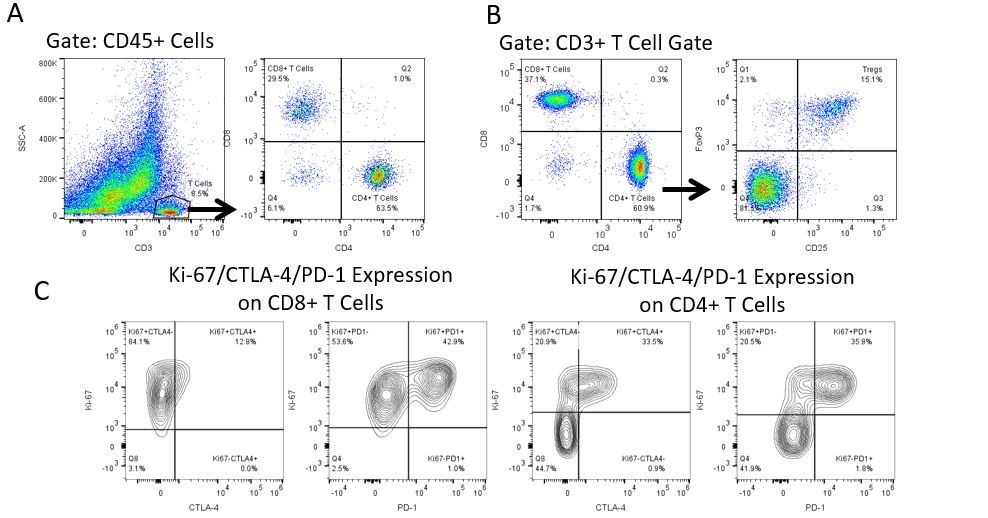
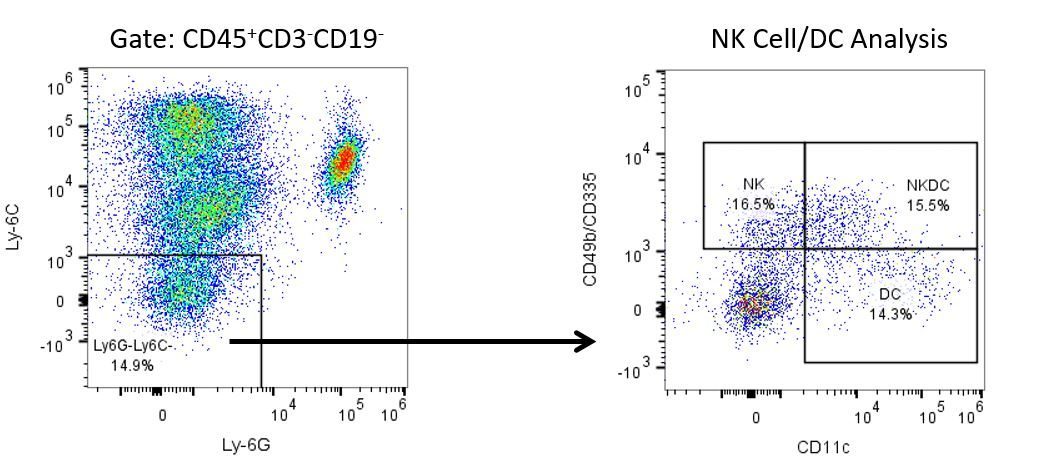
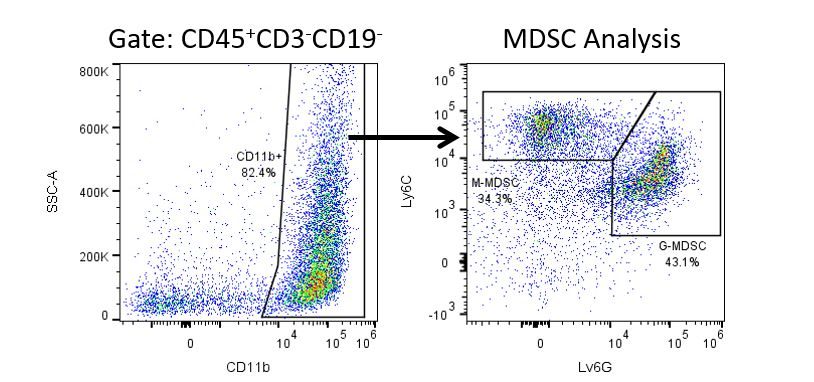
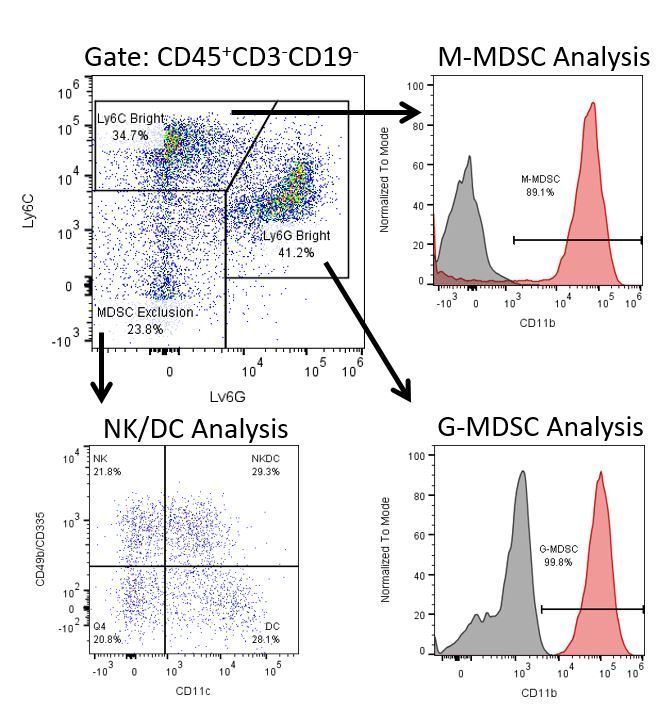
The most recent breakthroughs in cancer research and treatment have been in the field of immuno-oncology, using novel immunotherapies to bolster the host’s immune system to more effectively target and destroy tumor cells. Among the list of FDA-approved therapeutics with immunoregulatory activity are antibody and cytokine-based immunotherapies, small molecule drugs, and cell-based therapy. The success that these and other therapies have had at prolonging survival has placed emphasis on the development of new single agent and combination therapies with greater efficacy at treating cancer.
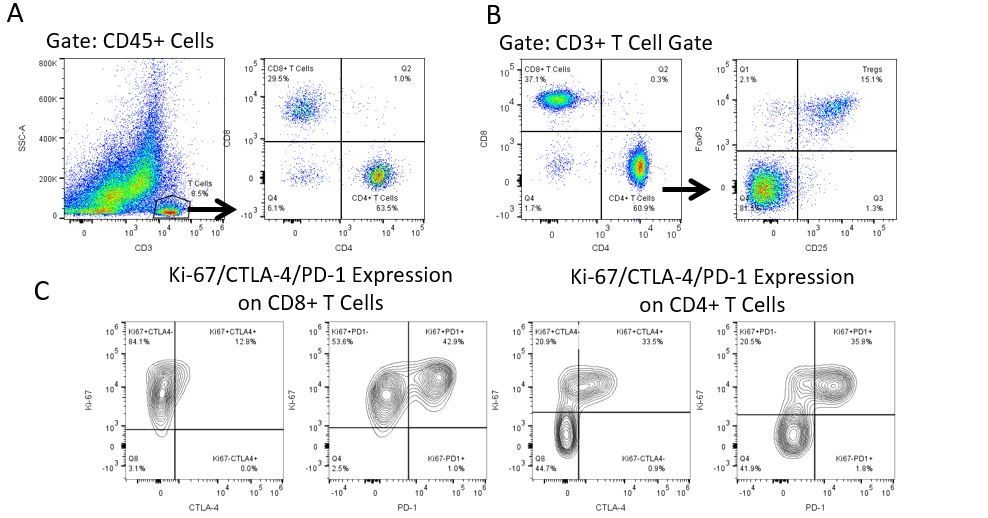
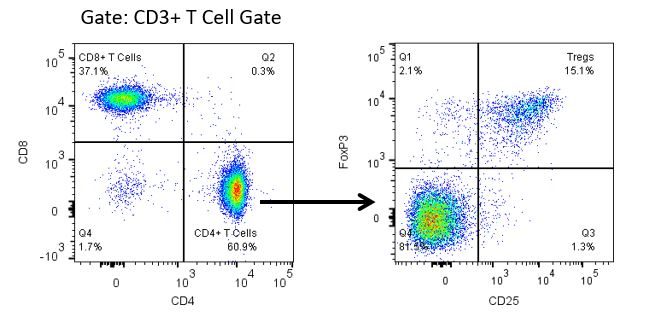
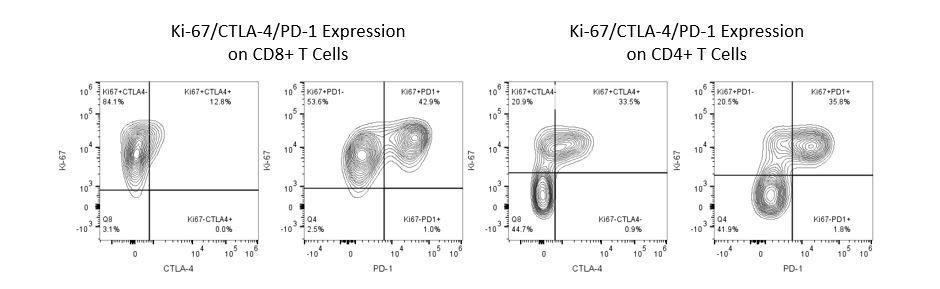
Productive development of immunotherapies requires high-throughput and robust quantitative analysis of the immune system in the tumor micro-environment, peripheral blood, and other host tissues.
To meet this need, our contract flow cytometry service, provides you with an advanced, state-of-the-art analytical flow cytometry resource to support all aspects of your drug development needs. Run your sample generation studies with us or overnight-ship us your preclinical or non-CLIA regulated clinical samples and we’ll take care of the flow cytometry for you.